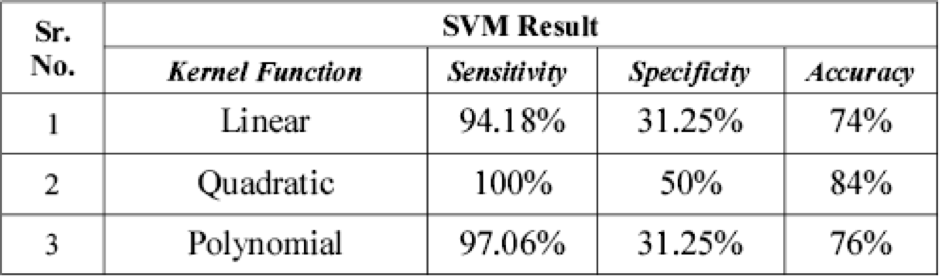
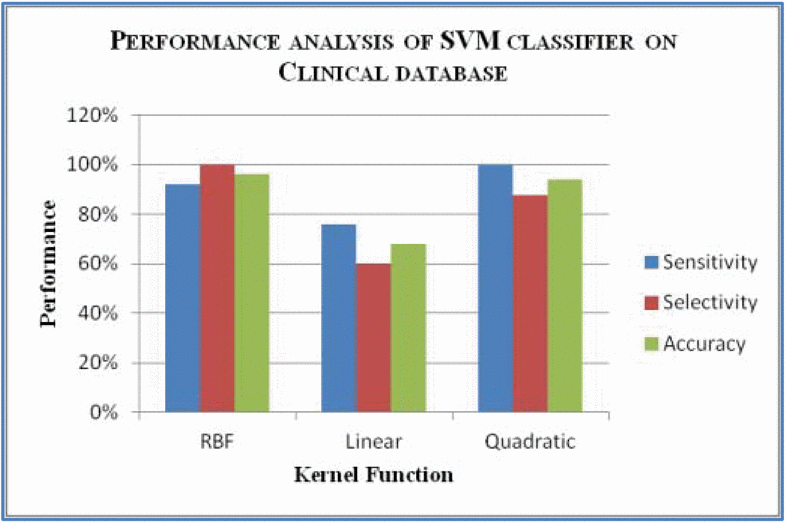
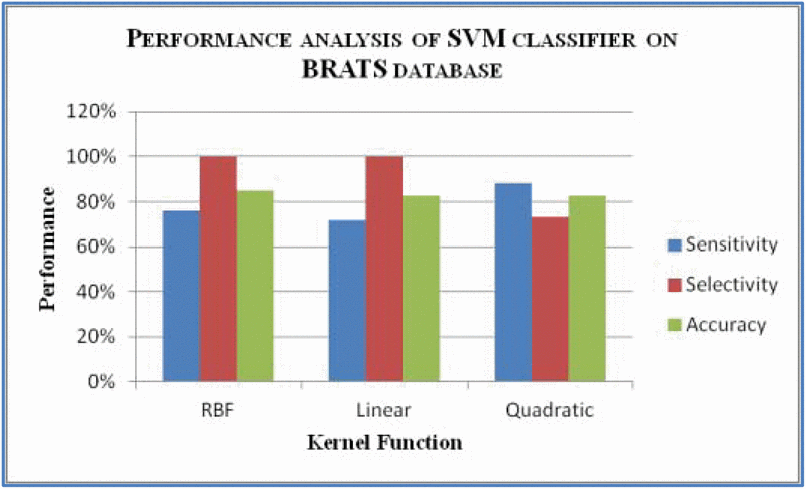
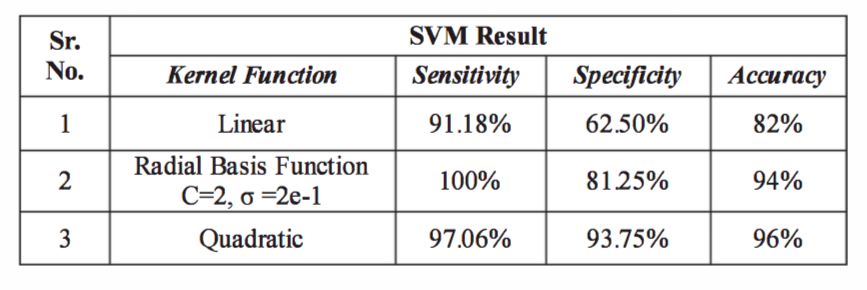
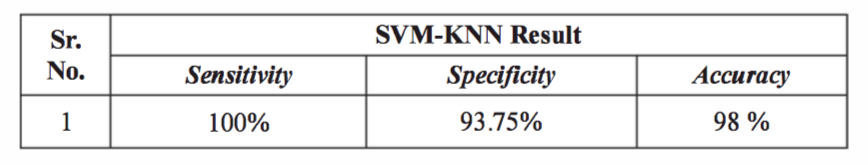
How do we test and measure performance of an SVM?
Given a trained SVM testing involves 'plotting' unseen data points (MRI images) in the space, and seeing if the SVM's classification of the data point matches the correct classification.
The options when classifying any data point, for the problem of classifying brain tumours as benign and malignant, are as follows:
- True positive (TP): Malignant brain tumour correctly identified as malignant
- False positive (FP): Benign brain tumour image incorrectly identified as malignant
- True negative (TN): Benign brain tumour image correctly identified as benign
- False negative (FN): Malignant brain tumour incorrectly identified as benign
The following statistical measures can be taken:
- Sensitivity: TP/(TP + FN) * 100%
- Specificity: TN/(TN + FP) * 100%
- Accuracy: (TP + TN)/(TP + TN + FP + FN) * 100%
Sensitivity can be thought of as the proportion of malignant brain tumours correctly identified, and specificity as the proportion of benign brain tumours correctly identified.
One can argue that sensitivity is more important than specificity since it is time critical to identify malignant tumours.